





 s with most imaging modalities, MR imaging data are normally presented as two-dimensional gray-scale images. However, MR imaging is essentially a 3D method and can produce three-dimensional data sets of virtually any body organ [⇒ Aichner 1994].
s with most imaging modalities, MR imaging data are normally presented as two-dimensional gray-scale images. However, MR imaging is essentially a 3D method and can produce three-dimensional data sets of virtually any body organ [⇒ Aichner 1994].
The simplest way of visualizing such data sets is by letting radiologist flip through the set displayed as slices of 2D images, leaving it up to them to visualize the structures.
Whereas this approach is suitable for some purposes, like diagnosis, it is less suitable for other purposes, like surgical planning or radiotherapy planning.
Thus, there is a need for 3D visualization techniques.
By performing segmentation, surface- or volume-rendering techniques can be applied [⇒ Maintz 1998]. The advantage of surface-rendering techniques is that they are easy and fast to visualize and manipulate (by rotation, zooming, etc.).
Since a segmentation has been done, it is possible to manipulate the 3D data set by removing tissues, request volumes and sizes.
The disadvantage is that segmentation of the data is required before visualization can be performed, and that some information is lost in the segmentation step. An alternative technique is volume-rendering. Volume-rendering does not require segmentation.
However, the method requires more powerful computers to be fully interactive, and normally some interaction to visualize structures of interest (Figure 15-12) [⇒ Tiede 1994].
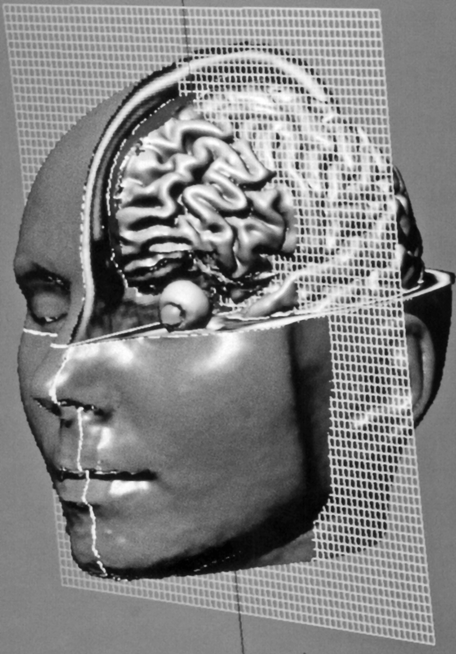
Figure 15-12:
Dissection of an MR imaging-based head model. A wire mesh is used to define cut planes. Similar reconstructions can be used in surgery and radiation therapy planning. One of the main problems in 3D image-processing is that objects within the 3D domain may obscure each other. Therefore any visualization must be preceded by a segmentation step in which 3D regions belonging to an organ must be identified.